Python: Analysing EMG signals – Part 1

Electromyography (EMG) is an experimental and clinical technique used to study and analyse electrical signals produced by muscles. This series of tutorials will go through how Python can be used to process and analyse EMG signals. We begin with a brief overview of how muscle electrical signals are produced and detected.
What is EMG?
The smallest functional unit that describes how a muscle contracts is the motor unit (Figure 1a). The motor unit consists of the cell body and axon of a motor neurone and the muscle fibres it innervates. Each muscle fibre is enclosed in a cell membrane surrounded by electrically charged particles (sodium and potassium ions). In a relaxed muscle fibre, there are more sodium ions outside the cell compared to inside it. The difference in concentration of ions sets up a potential difference of about -80 thousandths of a Volt (millivolt, mV) across the muscle cell membrane. (That is, there is a difference in charge or polarity across the membrane. You could think of the potential difference as an electrical “force” produced by charged sodium ions outside the cell “pushing” against the membrane to get in.) The imbalance in sodium ions (and consequently, the potential difference) is maintained by a cell membrane pump. When an electric current passes from the motor neurone to the muscle fibre, the current changes the chemical properties of the muscle cell membrane, causing sodium ions to pass from outside into the muscle cell which depolarises the cell membrane. The cell membrane pump then quickly restores the concentrations of sodium ions to conditions at rest, repolarising the membrane.
During depolarisation, if the change in potential difference reaches a certain threshold value (from -80 to -30 mV), an action potential is produced such that the cell membrane continues to depolarise even further to +30 mV before repolarising (Figure 1b). The action potential produces more chemical changes that cause the muscle fibre to contract, before the action potential travels down the muscle fibre. If a pair of electrodes is placed along the length of the muscle fibre, the electrodes can detect the big swing in potential difference during depolarisation and repolarisation (-80 to +30 mV) as the action potential passes between the electrodes while it travels along the muscle fibre. This is the mechanism by which muscle electrical signals can be detected.
Figure 1: Diagram of (a) a motor unit (Konrad 2006, Fig 5) and (b) an action potential (Konrad 2006, Fig 7).

When a motor neurone fires, the action potential travels from the axon to the muscle fibre and causes the fibre to contract (twitch), producing a tiny force. When many motor neurones fire, the action potentials produce tiny forces in many fibres, and the addition of all these forces (summation) produces a muscle contraction. Surface electrodes are usually used to detect the summation of action potentials from many motor neurones through the skin. Fine wire electrodes can be inserted into a muscle to detect the activity from several fibres near by that are part of the same motor unit – this method demonstrates the electrical discharge pattern of the motor neurone located in the spinal cord. Here, we will focus on processing and analysing muscle electrical signals from surface electrodes (surface EMG).
Simulation of muscle EMG
Let’s use Python to simulate some simplistic, non-physiological EMG data obtained from two maximal voluntary contractions of a muscle:
Figure 2: Simulated EMG data from 2 muscle contractions
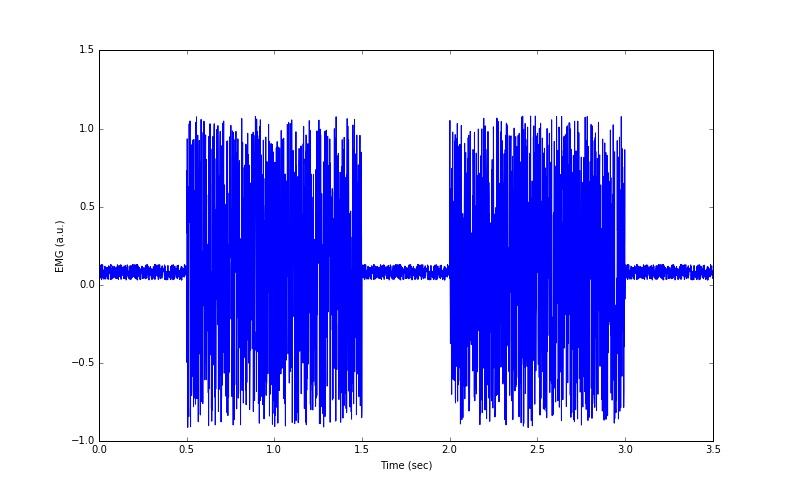
The code that generates Figure 2 is:
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
# simulate EMG signal
burst1 = np.random.uniform(-1, 1, size=1000) + 0.08
burst2 = np.random.uniform(-1, 1, size=1000) + 0.08
quiet = np.random.uniform(-0.05, 0.05, size=500) + 0.08
emg = np.concatenate([quiet, burst1, quiet, burst2, quiet])
time = np.array([i/1000 for i in range(0, len(emg), 1)]) # sampling rate 1000 Hz
# plot EMG signal
fig = plt.figure()
plt.plot(time, emg)
plt.xlabel('Time (sec)')
plt.ylabel('EMG (a.u.)')
fig_name = 'fig2.png'
fig.set_size_inches(w=11,h=7)
fig.savefig(fig_name)
Figure 2 shows simulated data of two EMG ‘bursts’ – the spikes are collections of action potentials from many neurones firing to stimulate muscle fibres. In the next part, we will discuss how the data were simulated, some of the problems with these EMG data, and what can be done to fix them.
Reference
Konrad P (2006) The ABC of EMG – A practical introduction to kinesiological electromyography. Noraxon USA Inc.

Hi! I just want to thank you for this fantastic tutorial! I’m a grad student training some undergrads and I always send them to your tutorial. Thanks for the great work!
LikeLike
Thank you for the feedback, much appreciated.
LikeLike
Hi Joanna
Thanks for this tutorial. I’d like to practice on the EMG signal that I’ve already acquired but I’m not sure how to load the signal into Python. Do you have any suggestions? THanks !
LikeLiked by 1 person
Hi there, importing data into Python depends on the file format the data are stored in. Python easily reads in text files: see these tutorials on importing text files into Python. If your data are in a binary format (eg. Spike files), see this tutorial for importing Spike2/Matlab files into Python. Cheers
LikeLiked by 1 person
Hi Joanna, Thanks so much – I’ll give it a try. 🙂
LikeLike
Hi Joanna, thanks for your answer and your help. I could get a solution about the problem with the graphic. The problem was that I had only a small part of the graphic from your post, but it was because I was running the code in Python 2, I confirmed the commentaries of the second EMG and I could solve the problem.
Thanks.
LikeLiked by 1 person
Hi Joanna, congratulations for your post, it is very interesting, so I have a detail, when I run the program to get the graphic of the Figure 2 I get a different graphic, help please.
LikeLike
Hi Andres, thanks for your comment, but I will need more details on how the figure you obtained was different from the one in the post.
I ran the code in this post and obtained a similar figure – similar but not identical, because
np.random.uniform()will generate different random numbers each time the function is called, so the simulated EMG spikes in your figure won’t be identical to the EMG spikes in Figure 2 in the post. This is the only minor difference I could find.The code was developed in Python 3 but if you happen to be running it in Python 2, take a look at the comments in response to a potentially similar problem in the second EMG post.
LikeLike